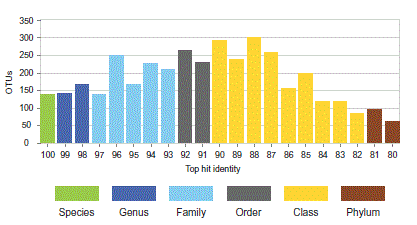
Top-hit identity distribution
See also
Cross-validation by
identity
usearch_global
command
A top-hit identity distribution (THID) is useful for
visualizing distances between a set of query sequences, e.g. OTUs, and a
reference database used to predict a trait, e.g. an RDP Classifier training
set. OTUs with low identities create a challenging problem for prediction
algorithms. Reviewing a THID for a set of OTUs gives an indication of
whether predictions based on a given reference are likely to be robust.
USEARCH does not currently support generating a THID as
an image file; if this would be useful for you,
please let me know. However,
it is straightforward to make one with the help of a small script.
Run the OTUs against the reference using the
usearch_global command, e.g.:
usearch -usearch_global otus.fa -db refdb.fa -strand plus
-id 0.5 \
-maxaccepts 8 -maxrejects 128 -top_hit_only \
-userout hits.txt -userfields query+target+id
Write a script to scan hits.txt. For each hit, round
identity to the nearest integer. Count the number of hits for each identity
value. Write the counts to a tabbed file and generate a histogram. The
example below is a THID for OTUs from a soil sample, colored to indicate the
most probable lowest common rank with the NCBI
BLAST 16S reference database.
References (please cite)
R.C. Edgar (2018), Accuracy of taxonomy prediction for 16S rRNA and fungal ITS sequences, PeerJ 6:e4652
• Cross-validation by identity, novel benchmark strategy enabling realistic accuracy estimates
• Genus accuracy of best methods is 50% on V4 sequences
• Recent algorithms do not improve on RDP Classifier or SINTAX
R.C. Edgar (2018), Taxonomy annotation and guide tree errors in 16S rRNA databases, PeerJ 6:e5030
• Approx. one in five SILVA and Greengenes taxonomy annotations are wrong
• SILVA and Greengenes trees have pervasive conflicts with type strain taxonomies