Taxa known only from sequence
See also
Microbial taxonomy
Defining microbial taxa
In microbial taxonomy, lower taxa, especially genera, are often defined by characteristic traits observed in type strains. However, a large majority of known 16S rRNA sequences were obtained from environmental samples and do not match sequences of isolate strains with known traits.
There are ~2,000 named genera in microbial taxonomic nomenclatures such as Bergey's Manual , LSPN and the NCBI Taxonomy database , while the diversity of sequences in large databases such as SILVA and Greengenes is roughly equivalent to ~100,000 genera, as shown in the table below from Yarza et al . 2014 . Thus, only ~2% of genera have been named and ~98% of known 16S sequences have unknown traits .
Therefore, most taxonomy annotations in large databases such as RDP, SILVA and Greengenes are not authoritative classifications based on observed traits , in fact they are predictions from sequence alone and many of these taxonomy annotations are wrong . See taxonomy annotation errors in large databases for details.
Yarza et al. used the CD-HIT clustering method to determine optimal identity thresholds corresponding to each rank, then estimated the number of taxa at each rank as the number of clusters at the optimal identity threshold. This approach has substantial methodological problems, including strong taxonomic bias which skews the thresholds (see Edgar 2018 , Accuracy of taxonomy prediction for 16S rRNA and fungal ITS sequences ) and also issues with CD-HIT including (a) a non-standard definition of sequence identity, and (b) low sensitivity which causes CD-HIT to report too many clusters by its own definition of identity.
At higher ranks, in particular phylum, I believe that Yarza et al. 's results greatly over-estimate the number of unnamed taxa due to the low sensitivity of CD-HIT, e.g. I doubt that only 27 / 1,481 = 2% of phyla in SILVA have been named. However, at genus rank the numbers are comparable to my own unpublished estimates obtained with UCLUST and corrections for bias, so I believe it is reasonable to estimate that ~ 2% of genera in SILVA have been named.
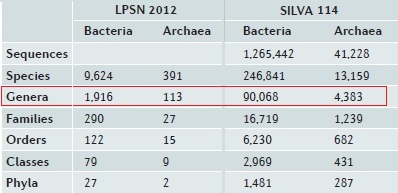
From Table 2, Yarza et al . 2014 .

