97% OTU threshold is too low
See also
Lowest common rank probabilities
calc_lcr_probs command
Setting OTU threshold
The canonical clustering threshold is 97% identity, which was proposed in 1994 when few 16S rRNA sequences were available, motivating a reassessment on current data.
Using a large set of high-quality 16S rRNA sequences from finished genomes, I assessed the correspondence of OTUs to species for five representative clustering algorithms using four accuracy metrics. All algorithms had comparable accuracy when tuned to a given metric.
Optimal identity thresholds were ~99% for full-length sequences and ~100% for the V4 hypervariable region.
This result is confirmed by calculating the lowest common rank probability for species as a function of identity, as shown in the figure below. This shows that at 97% identity, the probability that a pair of sequences is from the same species is almost zero, so it is more likely that they are from different species in the same genus, or even from different genera.
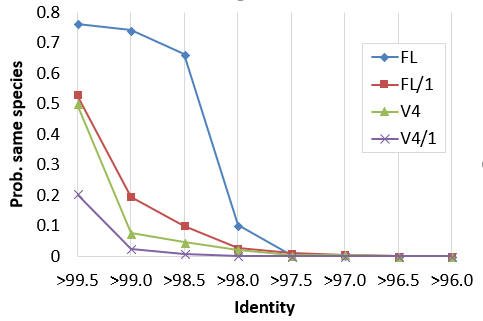
Probability that a pair of sequences is from the same species as a function of identity.
References (please cite)
R.C. Edgar (2017), Updating the 97% identity threshold for 16S ribosomal RNA OTUs, Bioinformatics 34(14) 2371-2375
• Standard 97% OTU identity threshold is too low
• Optimal OTU threshold is 99% for full-length 16S, 100% for V4
R.C. Edgar (2018), Accuracy of taxonomy prediction for 16S rRNA and fungal ITS sequences, PeerJ 6:e4652
• Cross-validation by identity, novel benchmark strategy enabling realistic accuracy estimates
• Genus accuracy of best methods is 50% on V4 sequences
• Recent algorithms do not improve on RDP Classifier or SINTAX
R.C. Edgar (2018), Taxonomy annotation and guide tree errors in 16S rRNA databases, PeerJ 6:e5030
• Approx. one in five SILVA and Greengenes taxonomy annotations are wrong
• SILVA and Greengenes trees have pervasive conflicts with type strain taxonomies

