Sequence databases with taxonomy annotations 
See also
Microbial taxonomy
The SILVA and Greengenes "taxonomies"
Taxonomic nomenclatures (naming systems)
Taxonomy errors in 16S databases
Conflicts between 16S trees and taxonomy
Specialized 16S rRNA sequence databases providing taxonomy annotations include Greengenes, LTP, RDP, and SILVA.
At the time of writing (mid-July 2018), it appears that Greengenes may be defunct as the last release was v13.5 from May 2014.
LTP (Living Tree Project) is based on sequences of type strains and isolates classified on the basis of observed traits. The other databases are much larger, containing mostly environmental sequences.
In the RDP database, taxonomy annotations of environmental sequences are predicted by the Naive Bayesian Classifier (NBC). I estimate the error rate of these annotations to be roughly 10%. A subset of type strain and isolate sequences is provided as training data for the Classifier.
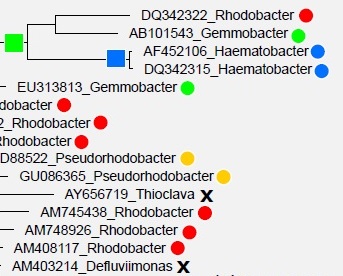 Greengenes and SILVA are annotated by a combination of database-specific computational prediction methods and manual curation based on predicted phylogenetic trees inferred from multiple sequence alignments. Roughly one in five of these annotations are wrong , almost certainly because of branching order errors in the trees .
Greengenes and SILVA are annotated by a combination of database-specific computational prediction methods and manual curation based on predicted phylogenetic trees inferred from multiple sequence alignments. Roughly one in five of these annotations are wrong , almost certainly because of branching order errors in the trees .

