16S trees conflict with each other and with type strain taxonomy
See also
Microbial taxonomy
Taxonomy annotation errors in large databases
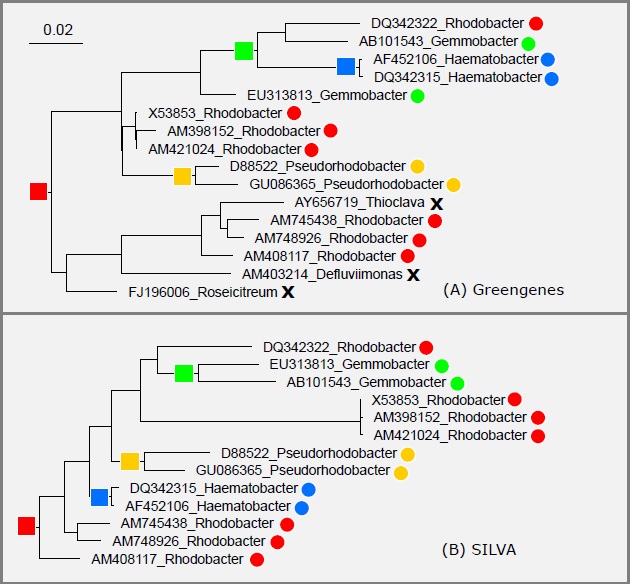
The guide trees in the SILVA, Greengenes and LTP databases have pervasive conflicts with each other and with type strain taxonomy. This is almost certainly explained by branching order errors in the trees compared with the correct tree for the 16S gene; see Edgar 2018 , Taxonomy annotation and guide tree errors in 16S rRNA databases for details.
As an example, the figure below shows the subtrees for genus Rhodobacter from SILVA and Greengenes. These subtrees have many conflicts with the type strain sequences at the leaves, meaning that most groups overlap and are therefore polyphyletic according to the tree. The SILVA and Greengenes trees also disagree with each other, so unless one tree is believed to be much better than the other, the most likely explanation is that both trees are wrong.
If one tree is more accurate than the other, then it should have better agreement with taxonomy defined by traits , i.e. should have more "pure" (monophyletic) genera, but my tests show that this is not the case. Therefore, both trees probably have comparable numbers of errors. See paper for details.
With the LTP tree, the subtree under the lowest common ancestor node for Rhodobacter contains 390 sequences from 122 genera. A figure for this this subtree is provided as as PDF file as it is too large to show on this page.