The SILVA and Greengenes "taxonomies" 
See also
Microbial taxonomy
Which taxonomy reference database should you use?
The literature describes a "Greengenes taxonomy" and "taxonomic frameworks" for SILVA and RDP (references below). I was confused about these for a long time, so I thought it would be helpful to clarify what these are and what they are not.
Contrast with the RDP database
Taxonomy annotations in the RDP database are easier to understand. For isolate and type strain sequences, they are authoritative classifications based on observed traits . Annotations of environmental sequences are predictions by the RDP Naive Bayesian Classifier . The nomenclature of the RDP taxonomy annotations is based on Bergey's Manual .
Most Greengenes and SILVA annotations are not authoritative classifications by taxonomists
It is a common misconception that the taxonomy annotations in these databases are obtained by expert microbiologists examining isolate strains. This is not the case -- as described below, most annotations are computational predictions for environmental sequences with unknown traits .
Errors in SILVA and Greengenes taxonomy annotations
Roughly one in five of the taxonomy annotations in SILVA and Greengenes are wrong , almost certainly because of branching order errors in the trees . I believe that the RDP annotations are more accurate, with roughly one in ten incorrect annotations.
Greengenes and SILVA define their own nomenclatures
Both databases define their own nomenclature . These nomenclatures agree on most well-established names, which are defined by traits . They differ with each other and from Bergey's mainly in revisions to resolve conflicts with sequence-based phylogenies and add new candidate groups identified in environmental sequences. If a taxon name is defined by traits , all nomenclatures implicitly agree on the traits. In other words, a taxon name such as Salmonella should, and does, mean the same thing in all nomenclatures where it appears. However, if a taxon name is defined as a group of lower taxa, there may be disagreements on which taxa are included due to the difficulty of inferring phylogenetic relationships from sequence .
Greengenes and SILVA do not introduce new taxon names
Names in the nomenclatures are taken from the literature. Greengenes and SILVA do not introduce new names as such, though they do invent their own combined names for groups which are believed to overlap such as Escherichia-Shigella and Rhizobium/Agrobacterium group .
Greengenes and SILVA use tree-based algorithms to predict taxonomy for environmental sequences
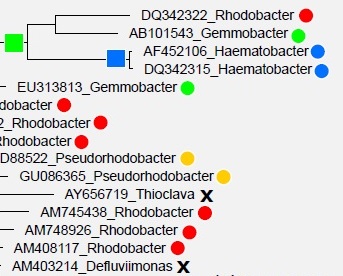 Most sequences in the databases are derived from environmental sequencing and have unknown traits . Both databases construct a multiple alignment which is used to estimate a tree, but the details differ. Taxonomy is predicted for environmental sequences using tree-based algorithms in an attempt to achieve consistency between phylogeny and taxonomy . The papers describe manual review and adjustments of the automated classifications, but to the best of my knowledge the databases do not document which adjustments are made or describe in detail why and how they are made. I would guess that most of the automated predictions are preserved (if nothing else, it would be too time-consuming to review hundreds of thousands of predictions manually), and the manual review in based
on placement in the predicted tree, though it is hard to imagine that the curators can make much sense of this because the tree topologies are not consistent with type strain taxonomies .
Most sequences in the databases are derived from environmental sequencing and have unknown traits . Both databases construct a multiple alignment which is used to estimate a tree, but the details differ. Taxonomy is predicted for environmental sequences using tree-based algorithms in an attempt to achieve consistency between phylogeny and taxonomy . The papers describe manual review and adjustments of the automated classifications, but to the best of my knowledge the databases do not document which adjustments are made or describe in detail why and how they are made. I would guess that most of the automated predictions are preserved (if nothing else, it would be too time-consuming to review hundreds of thousands of predictions manually), and the manual review in based
on placement in the predicted tree, though it is hard to imagine that the curators can make much sense of this because the tree topologies are not consistent with type strain taxonomies .
References
D. McDonald D et al . 2012, An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea, ISMEJ 6(3):610-618 https://doi.org/10.1038/ismej.2011.139 .
Yilmaz P et al . 2014. The SILVA and 'all-species Living Tree Project (LTP)' taxonomic frameworks , NAR 42(D1):D643-D648 DOI 10.1093/nar/gkt1209 .

