How to run a 16S sequence at NCBI BLAST
The NCBI BLASTN (nucleotide) web page is here:
https://blast.ncbi.nlm.nih.gov/Blast.cgi
Click "Nucleotide BLAST", which should give you a search form for "blastn suite".
Copy and paste your sequence in FASTA format into the text box at the top of the page.
Most of the default settings are fine, but you should usually change the database to "16S ribosomal RNA":
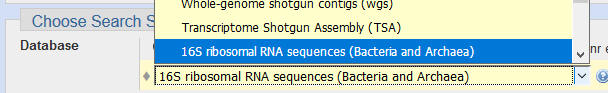
If this doesn't give a good hit, you can select "nucleotide collection nr/nt" which includes nucleotide sequences from the whole tree of life. This can be useful for identifying other sequences which are amplified due to unintended PCR primer matches.
, then scroll to the bottom of the page and click the "BLAST" button.