OTU accuracy results
See also
OTU benchmark
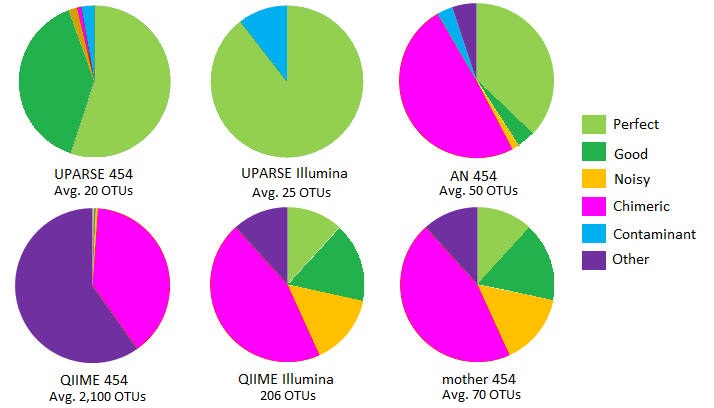
OTUs on mock community with 21 species .
Some species are missing from some samples; in those cases it is reasonable for a method to report < 21 OTUs.
In all 16S mock datasets , a large majority of UPARSE OTUs are Perfect, Good or Contaminant, i.e. are accurate reconstructions of biological sequences. See OTU classification for details of categories Perfect, Good etc.
By contrast, from 41% (Stag3P) to 71% (Even1P) of mothur OTUs were classified as Chimeric. A large fraction of QIIME OTUs was Chimeric, ranging from 23% (Stag3P) to 67% (Even1P). QIIME reported much larger numbers of OTUs than UPARSE or mothur on the pyrosequenced sets, ranging from 1,900 (Stag3P) to 3,647 (Stag2P). With AmpliconNoise, from 15% (Stag1P) to 64% (Even1P) of OTUs were chimeric.
Fungal mock communities
(Not shown in pie charts). On the fungal ITS communities, QIIME again produced large numbers of OTUs, none of which was within 3% of a known biological sequence, while UPARSE successfully reconstructed many of the expected species and many contaminants, though results on these datasets are harder to interpret. See the UPARSE paper for further discussion.
Reference
Edgar, R.C. (2013) UPARSE: Highly accurate OTU sequences from microbial amplicon reads, Nature Methods [ Pubmed:23955772 , dx.doi.org/10.1038/nmeth.2604 ].

