Fake chimeras
See also
Chimeras
Low-divergence chimeras are common
Given a query sequence Q, a chimeric model is a pair of reference sequence segments (A, B) concatenated together that have divergence > 0, i.e. are more similar to Q than the most similar sequence in the database (the top hit, T). If Q is not chimeric, the model is fake . A perfect fake model is a fake model that is identical to Q.
To give an indication of the frequency of fake models found by UCHIME2 I counted the number of alignments with a positive score, ignoring all other thresholds. Results are shown in the "Fakes" column in the table below (taken from the UCHIME2 paper). These are underestimates of the number of fake models that could be constructed, noting for example that this method fails to find many perfect fake models with S=99% found by UCHIME2-denoised.
All queries with at least one perfect fake model are guaranteed to be identified by UCHIME2 in denoised mode because the algorithm is not heuristic, in the following sense: if one or more chimeric models exist, and the query sequence is not present in the database, then a model will be reported.
The observation that both fakes and perfect fakes are very common implies that chimeras cannot be reliably distinguished from non-chimeras by any conceivable reference-based algorithm. In the de novo case, sequence abundances provide additional evidence which is predictive but not definitive. Similarly, it is not possible to screen large reference databases such as SILVA or Greengenes for chimeras with high accuracy. Low-divergence chimeras are common , and if the parents of a low-divergence chimera are present in the database, an algorithm such as UCHIME2-denoised can discover the model, but would not be able to reliably determine whether the model was fake because there is no evidence either way in the sequence. If its parents are not present, then the sensitivity and/or error rate of all algorithms necessarily increase rapidly with decreasing S and again, if a model is found, a chimera cannot be reliably distinguished from a correct biological
sequence with a perfect or imperfect fake model.
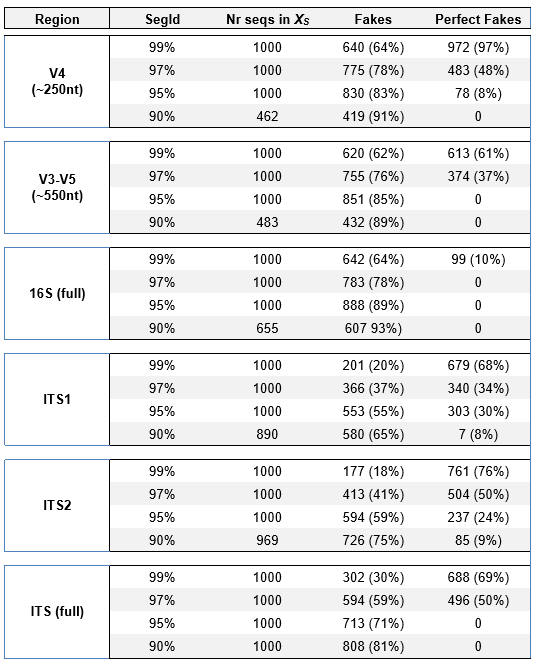
Table shows the number of fake models found by UCHIME2-sensitive and number of perfect fake models found by UCHIME2-denoised. For all regions except full-length 16S, a large majority of sequences have a perfect fake model with segment id (S) = 99% and between a third and a half with S=97%.
Reference (please cite)
R.C. Edgar (2016), UCHIME2: improved chimera prediction for amplicon sequencing, https://doi.org/10.1101/074252
• UCHIME2 algorithm, improved chimera detection
• "Fake" chimeras are common, valid biological sequences matching two-parent model
• Perfect chimera filtering impossible even with complete and correct reference
• Realistic chimera benchmark

