Cross-talk
 See also
See also
UNCROSS2 algorithm
UNCROSS paper
uncross command
Cross-talk example (GAIIx)
Cross-talk example (MiSeq)
Cross-talk errors assign reads to incorrect samples
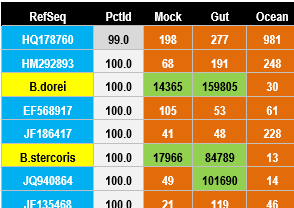
In marker gene amplicon sequencing, samples are often multiplexed into a single run by embedding index sequences into amplicons to identify the sample of origin. Reads are assigned to samples (demultiplexed) according to their index sequences. A cross-talk error occurs when a read is assigned to an incorrect sample.
Illumina has a ~0.1% cross-talk error rate
The cross-talk error rate was estimated to be ~0.1% in twelve Illumina datasets including one single-indexed GAIIx run and eleven dual-indexed MiSeq runs , as described in the UNCROSS paper . This means that on average, 0.1% of reads are assigned to the wrong sample. However, there are fluctuations, so in some OTUs the rate can be substantially higher. In a given OTU, the number of reads assigned to a single sample could be inflated by up to ~1% of the total reads in that OTU. Thus, if the OTU table shows that up to around 1% of the reads were assigned to a given sample, the correct count could be zero and this would then give a false-positive identification of the species (or group of species) in the OTU. Cross-talk thus tends to inflate estimates of richness and alpha diversity . Beta diversity may also
be inflated because samples may appear to share the same spurious OTUs.
Other next-generation machines seem to generate similar cross-talk rates. The reasons are unclear, though at least some cross-talk is probably due to read errors in the index (also called tag or barcode) sequences.
Control samples
Cross-talk can be identified most reliably in control samples such as a null sample (e.g. distilled water) and designed (mock) communities where the sequences are known.
Identifing and filtering cross-talk
The otutab_xtalk command attempts to identify and filter cross-talk in an OTU table.

