otutab_xtalk command
 Identify and filter cross-talk in an OTU table using the UNCROSS2 algorithm .
Identify and filter cross-talk in an OTU table using the UNCROSS2 algorithm .
The -minxtscore option specifies the score threshold. Default 0.1.
If there are mock community control samples, you can use the mockhits option to specify a tabbed text file reporting hits of OTU sequences to the mock community reference database. This enables more reliable identification of cross-talk into mock samples. The tabbed file should have three fields: #1. OTU label, #2. reference label, #3. percent identity. This format can generated by using the following options of the usearch_global command : -userout mockhits.txt -userfields query+target+pctid.
The -report option specifies a text file name for a report.
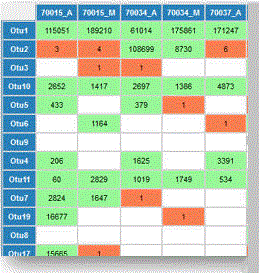
The -htmlout option specifies an HTML file name. This generates a web page for the OTU table where entries are colored according to their cross-talk score (see example on right).
The -otutabout option specifies the name of an OTU table output file in QIIME classic format . Counts with a score at or above the minxtscore threshold are set to zero.
Example
usearch -otutab_xtalk otutab.txt -report xtalk_report.txt \
-htmlout xtalk.html -otutabout otutabx.txt
Reference (please cite)
R.C. Edgar (2018), UNCROSS2: identification of cross-talk in 16S rRNA OTU tables, https://doi.org/10.1101/400762
• Cross-talk rate is approx. 1% in many Illumina datasets
• Cross-talk can cause false positive core microbiome
• UNCROSS2 algorithm for filtering cross-talk

