See also
UNBIAS algorithm
unbias command
 Read
abundance does not correlate with species abundance
Read
abundance does not correlate with species abundance
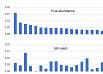
Using mock
community sample with species abundances determined independently by shotgun
sequencing, I found that 16S amplicon read frequencies have no meaningful
correlation with species frequencies (Pearson coefficient r close
to zero). Click here for figure.
The factors described below cause read frequencies to diverge substantially
from species frequencies.
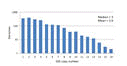 Gene copy number
Gene copy number
Prokaryotic genomes contain varying
numbers of 16S genes ranging from one to ten or more, and strains with more
genes therefore tend to be more common in the reads.
Click here for figure.
Primer mismatches
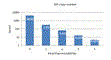 PCR amplification efficiency is
strongly degraded if a template has mismatches with the primers, causing the
number of reads to be suppressed, typically by an order of magnitude or more
for each mismatched position. With the currently popular V4 primers, ~9% of
species have one or more mismatches. Click
here for figure.
PCR amplification efficiency is
strongly degraded if a template has mismatches with the primers, causing the
number of reads to be suppressed, typically by an order of magnitude or more
for each mismatched position. With the currently popular V4 primers, ~9% of
species have one or more mismatches. Click
here for figure.
Sequence composition
GC content and homopolymers affect polymerase
efficiency.
Sequence length
Shorter sequences amplify more efficiently.
Currently popular 16S tags such as V4 have well-conserved lengths, but other
markers such as fungal ITS are more variable and therefore have stronger
amplification biases.
Degenerate primers
When degenerate primers are used, as
is commonly the case in 16S sequencing, biases occur due to unevenness in
the oligonucleotide mixture.
Biases are amplified
Small biases in efficiency, e.g.
due to uneven mixing of oligos, are exponentially
amplified by the PCR reaction, leading to large biases in read counts.
For example, if one sequence is amplified 10% more than
another in one round, it will be 1.120 = 7 times more abundant
after 20 rounds.