See also
Making an OTU
table
Downstream analysis with
QIIME
QIIME classic format is a tab-separated text used to store an OTU table. Newer versions of QIIME are moving to BIOM format for OTU tables, though in QIIME v1.9 some (many?) scripts still support classic files.
The first line has column headings, the remaining lines are OTUs.
The first column heading is "#OTU ID" (the quotes are not included, and there is exactly one space between OTU and ID). The other headings are sample names.
Optionally, the last column is used for a taxonomy annotation. I'm not sure which QIIME scripts use / need this column, or what the formatting requirements for the taxonomy names and ranks. If you need taxonomy in a classic table and run into problems, please let me know and I'll update this documentation and add features to USEARCH as needed.
A value in the matrix is an integer count, i.e. the number of reads for that OTU in that sample. As far as I know, fractional values such as frequencies are not supported by QIIME. It is up to the user to keep track of which type of count is used, e.g. raw, normalized, subsampled or rarified.
WARNING -- QIIME doesn't like underscores in OTU names
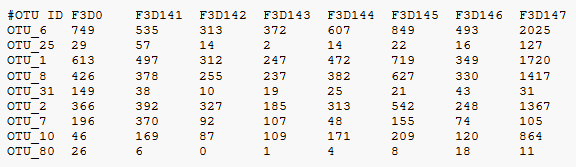
Some of my examples use OTU idenfiers like OTU_123, as in the table below.
Underscores in OTU idenfiers can cause problems with QIIME, apparently
because the
Newick tree file standard uses underscore to mean a blank space (because
the problem only seems to occur when a tree file is used). Some USEARCH
commands only allows letters, digits and underscores in OTU identifiers, so
you can't use another punctuation symbol (e.g., a period). The safest choice
is to avoid underscores and use something like Otu123.
Example QIIME classic OTU table