See also
Making an OTU
table
Downstream analysis with
mothur
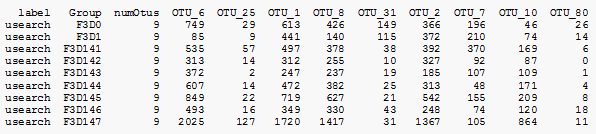
The mothur "shared" file is an OTU table.
The file is tab-separated text. The first line has column headings, the remaining lines are samples.
The first column is called "label". Most mothur commands ignore it. Some mothur commands set the label field to the clustering threshold, e.g. 0.03 for 97% OTUs. USEARCH commands put "usearch" in the label field.
The second column is called "Group". This is the sample name.
The third column is "numOtus". This is set to the number of OTUs, which is the same for all samples. It is redundant (because it can be calculated by counting the columns and subtracting three). If you modify a shared file to add or delete OTUs, you must be careful to update numOtus, so for example you can't use the Linux cut command to extract a subset.
A value in the matrix is a count, i.e. the number of reads for that OTU in that sample. Frequencies are not supported (because values must be integers). I'm not sure if / how mothur tracks whether the numbers are raw, normalized, subsampled or rarified. Possibly this is done through a file naming convention as this appears to be a common technique used in mothur, though the details are not documented and I find them to be obscure / incomprehensible. Here is a fairly typical shared filename taken from the mothur MiSeq SOP:
stability.trim.contigs.good.unique.good.filter.unique.precluster.pick.pick.pick.an.unique_list.shared
Example shared file