Random forest classifier
 See also
See also
Machine learning in OTU analysis
otutab_forest_train command
forest_train command
forest_classify command
Wikipedia page describing random forests
K-fold cross-validation
OTU importance
A random forest classifier is a machine learning algorithm for automatically predicting the category of an observation.
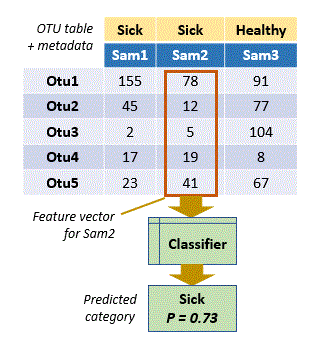
An observation is represented as a vector of numerical values, one for each feature of the observation.
In OTU analysis, an observation is a sample, a feature is an OTU, and the numerical value of the feature is the count or frequency of the OTU in that sample. Thus, a feature vector is a vector of OTU counts or frequencies for a sample. In this context, each column (sample) in an OTU table is a feature vector.
The category is an attribute of the sample, e.g. healthy or sick, high or low acidity, etc. In microbiology, attributes are often called "metadata", and a category (e.g. healthy) is called a "metadata state".
The parameters of a classifier are trained on observations (samples) which are labeled with the correct category. The resulting classifier can then be used to predict categories of novel samples. However, this approach is rarely used in practice in OTU analysis.
The most common use of machine learning classifiers in OTU analysis is to answer the question: does the composition of the community change with the sample metadata state (healthy / sick etc.)? Put another way, can metadata states be predicted from OTU counts or frequencies? Random forests are an effective way to address this question because they are very flexible and robust. For example, unlike most other classifier algorithms, they are highly tolerant of many uninformative features (i.e., OTUs which do not correlate with sample attributes), and make few assumptions about the structure of the data.
Random forests are based on discovering rules which are constructed from Boolean expressions with fixed numerical thresholds for the counts or frequencies of given OTUs. For example:
Sample is "Healthy" if (OTU64 has count>100 and OTU12 has count<10) or (OTU4 has count=0).
Sample is "Sick" if (OTU10 has count>1 and OTU11 has count>1 and OTU15 has count>2).
Important OTUs , i.e. those which are predictive of metadata states (i.e., are used most often in the rules discovered by the classifier) can be identified in the forest parameter file .
I usually don't recommend trying to identity informative OTUs by training a random forest on the complete OTU table as this may tend to over-tune the parameters. It is better to use k-fold cross-validation . Usually, OTUs are informative because their abundances correlate with metadata state, e.g. an OTU frequency might be tend to be high in a sick person and low in a healthy person. It is simpler and more direct to use the otutab_select command to identify those OTUs. Comparing results from otutab_forest_kfold and otutab_select is useful to check whether the rules discovered by the random forest are mostly simple correlations (these OTUs will be identified by otutab_select) or have more complex associations (which will not be discovered by otutab_select).
Reference
L. Breiman (2001) Random Forests , Machine Learning. 45 (1): 5-32. doi: 10.1023/A:1010933404324

