FAQ: Running cluster_fast/smallmem on metagenomics contigs
UCLUST is not designed for shotgun contigs
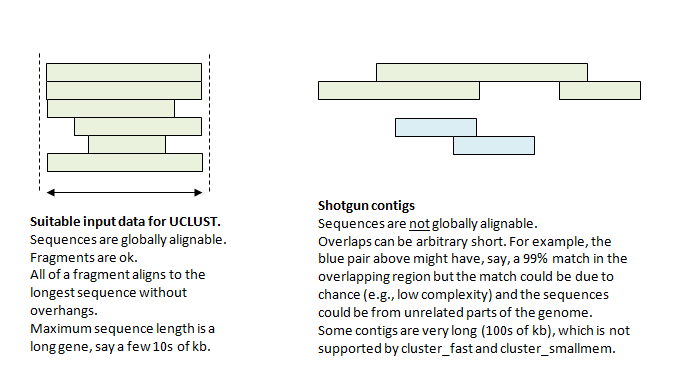
The UCLUST algorithm assumes that the input sequences belong to groups that are globally alignable, such as genes. Shotgun reads and contigs are not suitable input, as shown in the figure below
Metagenomic assembly algorithms are designed to find overlaps in shotgun reads and combine them into contigs. After assembly, there should be no remaining overlaps that can be reliably detected to belong to contiguous sequences. It therefore does not make conceptual sense to use an general-purpose sequence clustering algorithm like UCLUST on this data.
You could find ORFs then cluster the ORFs by using UCLUST and/or by aligning ORFs to known genes, e.g. using ublast against a large protein database, or by idenifying protein domains using Pfam .