otutab_trim command
See also
Making an OTU table (otutab command)
Setting an OTU abundance cutoff using an octave plot
Remove low-abundance counts, samples and OTUs from an OTU table . The input file must be in QIIME classic format . The output file is specified by the -output option. It is written in QIIME classic format.
Options to set minimum abundances are min_sample_size, min_count, min_freq, min_otu_size and min_otu_freq. See below for definitions and defaults. 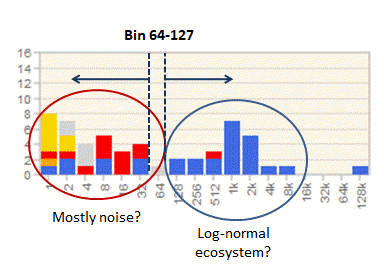 .
.
Determining an OTU abundance cutoff
See setting an OTU abundance cutoff using an octave plot .
Deleting samples with too few reads
Setting a minimum number of reads for a sample can be quite subjective and depends on the objectives of your analysis. As a rule of thumb, I prefer to have at least 5,000 reads per sample. Example command line:
usearch -otutab_trim otutable.txt -min_sample_size 5000 -output trimmed.txt
Options which specify minimum abundances
-min_sample_size.
Default 1. Samples with total count less than this are deleted.
-min_count
Default 1. Minimum count. Counts less than this are set to zero.
-min_freq
Default 0.0. Minimum frequency, defined as count divided by total sample size. Counts smaller than this are set to zero.
-min_otu_size
Default 1. Minimum total size for an OTU. OTUs smaller than this are deleted.
-min_otu_freq
Default 0.0. Minimum size for an OTU as a fraction of all OTUs. OTUs smaller than this are deleted.
If no minimums are specified, the effect is to delete samples and OTUs which have zero.total size.

