Chimera detection benchmarks
See alsoOTU benchmark home
Algorithm comparisons
UCHIME2 benchmark
UCHIME vs. ChimeraSlayer
UCHIME vs. Perseus (AmpliconNoise)
UCHIME vs. DECIPHER
UPARSE-OTU (cluster_otus) vs. UCHIME
Note
The test described in the ChimeraSlayer paper and used in the first UCHIME paper is misleading because it does not count false negatives as errors, but false negatives are equally important to false positives. The benchmark tests described in the UCHIME2 paper are more realistic.
Conclusions
The UPARSE-OTU algorithm (implemented in the cluster_otus command ) is currently (mid-2018) the most effective chimera filter for 97% OTU clustering.
The UCHIME2-denoised-denovo algorithm used by UNOISE3 (as implemented in the unoise3 command ) is superior to the DADA2 chimera filterring strategy and is currently (mid-2018) the most effective chimera filter for denoising (see results and discussion in UNOISE2 paper).
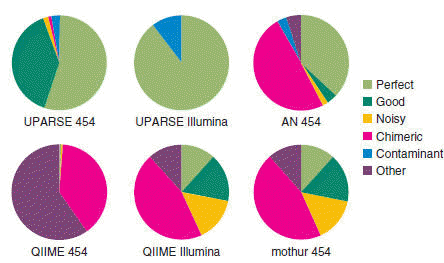
Fig 1(a) from the UPARSE paper . Magenta segments in the pie chart show the fraction of chimeric OTUs generated by several algorithms on a mock community test. The QIIME and mothur pipelines used UCHIME for chimera detection. Notice that even after UCHIME filtering, about 50% of the OTUs generated by QIIME and mothur are chimeric, while UPARSE successfully filters all chimeras from Illumina reads and leaves only one chimeric OTU with 454 reads.
References (please cite)
R.C. Edgar et al. (2014), UCHIME improves sensitivity and speed of chimera detection, Bioinformatics 27(16) 2194-2200
• Shows UCHIME faster and more accurate than ChimeraSlayer
• This paper report misleading benchmark tests, see critique in UCHIME2 paper
R.C. Edgar (2016), UCHIME2: improved chimera prediction for amplicon sequencing, https://doi.org/10.1101/074252
• UCHIME2 algorithm, improved chimera detection
• "Fake" chimeras are common, valid biological sequences matching two-parent model
• Perfect chimera filtering impossible even with complete and correct reference
• Realistic chimera benchmark
R.C. Edgar (2016), UNOISE2: improved error-correction for Illumina 16S and ITS amplicon sequencing, https://doi.org/10.1101/081257
• UNOISE2 algorithm, improved denoiser
• Reduces false-positive chimeras compared to UNOISE and DADA2

