|
See also
Search flowchart
The USEARCH algorithm searches a database for high-identity hits to
one or more database sequences ("targets"). USEARCH is used by the
usearch_global and usearch_local commands and is used as a
subroutine by cluster_fast and
cluister_smallmem. This
algorithm is fundamentally different from the UBLAST algorithm that is designed for low
identity local searches.
Recommended identity
ranges
USEARCH is effective at identities of ~50% and above for proteins and ~75%
and above for nucleotides.
Word counts and
U-sorting
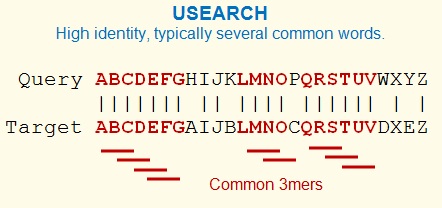
USEARCH exploits that fact that similar sequences
tend to have several short words in common. These words have a
fixed length k, and are sometimes called kmers. Unlike other
programs that use kmer counting, USEARCH does not attempt to
estimate sequence identity from the number of matching kmers. This
is because identity correlates only approximately with the word
count and does not give an accurate estimate, especially for lower
identities. Instead, USEARCH uses the word count to prioritize the
database search. Target sequences are compared to the query in
order of decreasing unique word count (U). This is the "U" in
USEARCH. If a hit above the identity
threshold exists in the database, it is likely to be found
close to the start of the U-sorted list of targets (the "U
vector").
Accepts and
rejects
An "accept" is a target sequence that is above the
identity threshold so can be considered a hit. A "reject" is a
target sequence that is compared to the query but falls below the
threshold. If targets are compared to the query in U-sorted order,
then:
(i) the first hit that is
found is likely to be the best hit that exists in the database, or
close to it, and
(ii) the more rejects that
have occurred without a hit, the less likely it is that a hit
exists.
This means that the search
can be terminated early if (i) a hit is found, or (ii) several
rejects have occurred, with only a small loss in sensitivity. The
termination options
‑maxaccepts and ‑maxrejects determine when the search is stopped.
This technique can give a dramatic improvement in speed, which is a
decisive advantage for the very large sequence datasets that are
increasingly common in biology.
Word index
USEARCH
uses an index on words in the database for rapid calculation of the
U vector for a given query. Index
options allow fine tuning of speed, sensitivity and memory use.
The index can be stored in a UDB file,
which can be advantageous when a large database will be searched
repeatedly. Alternatively, the index can be constructed on the fly,
for example when a database is provided
in FASTA format. Indexes are also used by cluster_fast and cluister_smallmem, which use USEARCH as
a subroutine to match query sequences to a database of centroids of
existing clusters. With clustering, the index is necessarily
constructed on the fly since the database is updated as new
clusters are identified.
Reference
Edgar,
R.C. (2010) Search and clustering orders of magnitude faster than
BLAST, Bioinformatics 26(19), 2460-2461.
doi:
10.1093/bioinformatics/btq461
|
