 See also
See also
OTU / denoising pipeline
Alpha diversity
Beta diversity
Rarefaction
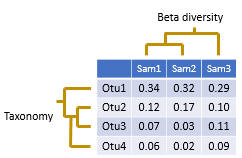
Diversity analysis investigates questions such as "how many species are in a sample?" and "how similar are these two samples?". The diversity in a single sample is called alpha diversity, and the diversity (differences or similarities) between two samples is called beta diversity.
The input data used for diversity analysis is an OTU table. A tree for the OTUs is also needed for UniFrac beta diversity analysis, this can be created using the cluster_agg command.
Usearch supports diversity analysis through the alpha_div, alpha_div_rare and beta_div commands. These commands support several popular diversity metrics.
Note that most standard diversity metrics are
difficult to interpret or invalid for
NGS OTUs.
Taxonomy for the OTU sequences can be predicted
using the sintax command.
A tree
for the OTU sequences can be generated using the
cluster_agg command.