See also
Read quality filtering
FASTQ format options
Quality scores
Global trimming
Python scripts
Amplicon reads are generated in many different formats depending on the sequencer type, base-caller software version, library preparation etc. The reads may include a combination of biological sequence (e.g., from the 16S gene) and non-biological sequences for calibration, sequencing (e.g. adapters) and sample identification (barcodes), and primer sequences.
Before analyzing your reads, you must first understand the "read layout", i.e. exactly how these different types of bases appear. Some typical examples are shown in the figures below; other variants are possible.
Prior to the dereplication step in OTU clustering, non-biological sequences must be removed. Usually, there is a barcode to identify the sample, though in some cases the sequencing machine software pipeline may have already split out your reads by barcode and removed the barcode sequences. To enable mapping of reads back to samples, the sample identifier can be added to the read label. In the case of 454, there may also be a calibration sequence, which is usually TCAG though sometimes the four letters appear in a different order. Sometimes the calibration sequence is removed by the base-calling software, so it may not appear in your FASTQ file.
Python scripts are provided to remove non-biological sequences and convert barcodes to read label annotations for typical read layouts. If your layout differs, you may need to edit the scripts or provide your own method of pre-processing the reads before quality filtering or dereplication.
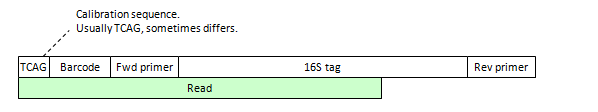
(A) 454 unpaired read, does not extend to second primer
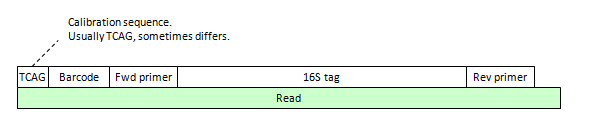
(B) 454 unpaired read, extends to second primer

(C) Illumina unpaired read, does not extend to second primer

(D) Illumina paired read, pair overlaps and covers both primers