DECIPHER and UCHIME compared
Validation of DECIPHER on published
benchmarks
DECIPHER is a 16S
chimera detection method
published in 2012.
The authors claim that DECIPHER has performance comparable to
UCHIME, especially with longer
sequences and when the parents of the chimera are at least 10% diverged
from each other. These claims are based mostly on results measured on
their own simulated chimeras. Method rankings on previously published
benchmarks are not reported in the paper. My results show that DECIPHER
in fact has substantially lower sensitivity than UCHIME, even in those
regimes where the authors claim DECIPHER has highest accuracy. It is not
clear to me why the performance of DECIPHER is better on the authors'
own test set; overtraining is one possible explanation.
Benchmark sets
I tested DECIPHER on the two of the benchmarks we used in the
UCHIME paper:
SIM2 and MOCK, both of which were developed by authors of previously
published chimera methods. SIM2 is a set of simulated chimeras from the
Haas et. al ChimeraSlayer paper. SIM2 has bimeras from 200nt to
full-length genes with zero to 5% added noise. MOCK contains denoised
reads from several mock community experiments in the Quince et al.
in the Perseus paper. These reads are inferred to be "good" (i.e., non-chimeric)
or to be chimeras by mapping reads to the known 16S sequences of the
individually sequenced type strain bacteria in these communities. Reads
of chimeric amplicons in these experiments are observed to have two,
three or four segments. The MOCK experiments used wet-bench sequencing
in which chimeras form naturally as a side effect of amplification,
avoiding the assumptions and simplifications required for in silico
simulation.
Results
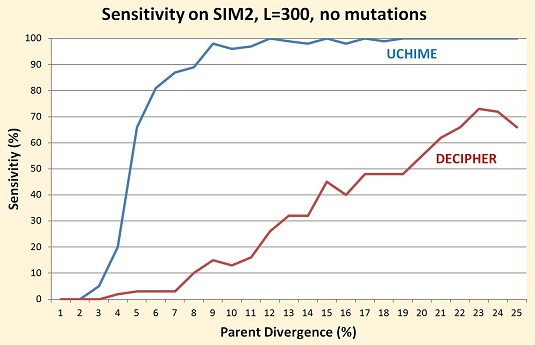
On all sets I've tested so far, the sensitivity of DECIPHER is
substantially lower than UCHIME, as shown in the table and figure below.
I tried to identify subsets that would support the authors' claim that
DECIPHER works best with longer sequences and when parent divergence is
high, but even in these cases DECIPHER has poor performance.
|
Benchmark and subset |
UCHIME |
DECIPHER |
|
MOCK Uneven, all chimeras |
94% | 63% |
|
MOCK Uneven, div ≥ 10% |
89% | 77% |
| SIM2, full-length, 2% indel noise, D ≥ 20% | 100% | 69% |
| SIM2, L=300, no mutations, D ≥ 10% | 99% | 46% |
| SIM2, L=300, no mutations, all chimeras | 81% | 31% |
Comment on Table 1 in the DECIPHER paper
In Table 1, the authors present DECIPHER results for SIM2 sets with
from 1 to 5% mutations. Only chimeras with at least 20% parent
divergence were included (the motivation for this choice is not
explained), and UCHIME results are not included. DECIPHER is reported to
have from 31% to 76% sensitivity, while UCHIME has 100% sensitivity on
all subsets.
